Chemlambda for the people
(no js version)
We can program a computer to do anything.
What if we had the same power over the molecules of our bodies?
Let's imagine how this could change our lives.
"still he'd see the matrix in his sleep, bright lattices of logic unfolding across that colorless void"
William Gibson, Neuromancer
"like making lists, just, fold up inside themselves. Come out the other way around. Crazy things."
Pseudo -- William Gibson

digital chemistry
to biological chemistry
and back
chemlambda
to biological chemistry
and back
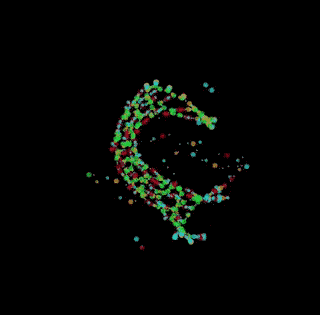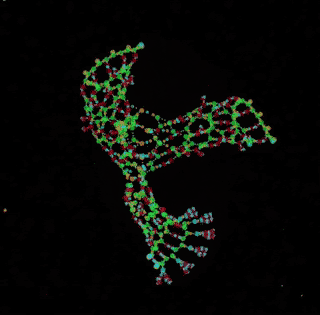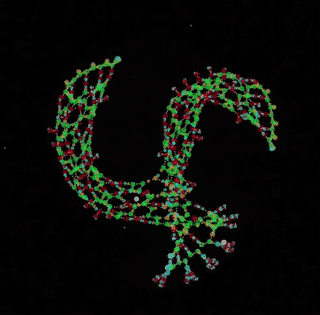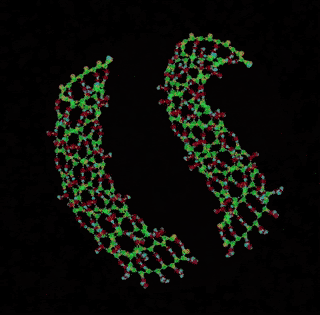

digital chemistry
to biological chemistry?
and back
Notes:
Continue to chemlambda projects page
